
The runtime performance of the proposed algorithm outperforms most of the existing algorithms for large network motifs. Further, the present work avoids the unnecessary growth of patterns that do not have any statistical significance. Current algorithms lack control of key network parameters, the ability to specify to what subgraphs a node belongs to, come at a considerable complexity cost or, critically and sample from a. The proposed algorithm can identify large network motifs up to size 15 by significantly reducing the computationally expensive subgraph isomorphism checks. In the EQA algorithm, BFS (Breadth First Search)-based candidate set generation strategy, matched vertices-based pruning strategy and query position-based. Designing algorithms that generate networks with a given degree sequence while varying both subgraph composition and distribution of subgraphs around nodes is an important but challenging research problem. This algorithm uses a pruning strategy to overcome the space limitation of the static expansion tree. The AGM algorithm employs a vertex-growth approach to generate a candidate (k+1)-subgraph (has k + 1 vertices) in level k + 1 by joining two frequent.

Motivated by the computational challenges of network motif discovery and considering the importance of this topic, an efficient and scalable network motif discovery algorithm based on induced subgraphs in a dynamic expansion tree is proposed. Finding network motifs in biological networks is a computationally hard problem due to their huge size and abrupt increase of search space with the increase of motif size. See core/model_utls/pyg_gnn_wrapper.py for more options.Ĭustom new GNN convolutional layer 'X' can be plugged in core/model_utls/pyg_gnn_wrapper.py, and use 'X' as model.gnn_type option.Biological networks are powerful representations of topological features in biological systems. core/model_utils/pyg_gnn_wrapper.py is the place to add your self-designed GNN layer X and then use X-AK(+) on fly~.SubgraphDrop is implemented inside core/transform.py, see here.Then for batch, all combined gaint subgraphs are combined again.) (For each graph with N node, N subgraphs are combined to a gaint subgraph first. In ith stage pick all subgraphs generated in (i-1)th stage, and extend the graphs by exactly one edge.

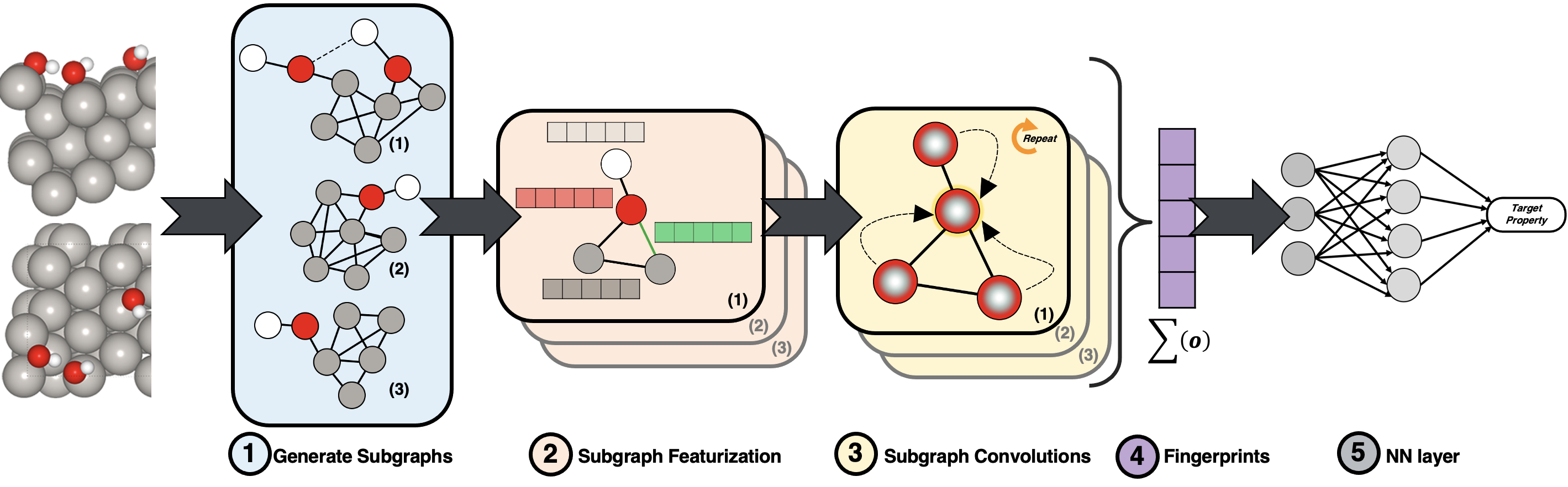
In the ith stage, generate all the subgraphs of the given graph that have exactly i edges.
#Subgraphs generator algorithm install
Pip install yacs=0.1.8 -force # PyG currently use 0.1.6 which doesn't support None argument. In this paper we describe a database containing 72,800 couples of simple graphs especially devised for comparing the performance of isomorphism and graph-subgraph isomorphism algorithms. Conda install pytorch=$TORCH torchvision torchaudio cudatoolkit=$cuda -c pytorch -c nvidia -yĬonda install pyg=$PYG -c pyg -c conda-forge -y


 0 kommentar(er)
0 kommentar(er)
